Shopping Cart
- Remove All
 Your shopping cart is currently empty
Your shopping cart is currently empty

Torin 1 is an effective inhibitor of mTORC1/2 with (IC50: 2 nM/10 nM); has 1000-fold selectivity for mTOR than PI3K.
| Pack Size | Price | Availability | Quantity |
|---|---|---|---|
| 1 mg | $30 | In Stock | |
| 2 mg | $41 | In Stock | |
| 5 mg | $67 | In Stock | |
| 10 mg | $97 | In Stock | |
| 25 mg | $183 | In Stock | |
| 50 mg | $328 | In Stock | |
| 100 mg | $496 | In Stock | |
| 500 mg | $1,070 | In Stock |
| Description | Torin 1 is an effective inhibitor of mTORC1/2 with (IC50: 2 nM/10 nM); has 1000-fold selectivity for mTOR than PI3K. |
| Targets&IC50 | mTORC2:10 nM, mTORC1:2 nM |
| In vitro | METHODS: Wild-type MEFs cells were treated with Torin 1 (250 nm) for 4 days, and cell proliferation was detected by CellTiterGlo viability assay. RESULTS: 250 nm Torin1 completely inhibited cell proliferation. [1] METHODS: The human endocrine cell line BON was treated with Torin 1 (62.5-150 µM) for 3-24 h, and the expression level of target proteins was measured by Western Blot. RESULTS: Torin1 increased NT secretion in a dose-dependent manner; NT secretion was increased 3-24 h after Torin1 treatment. Torin1 treatment decreased the phosphorylation of Akt (S473), and the phosphorylation of p70S6K (T389) and 4E-BP1 (T37/46) was also inhibited. [2] |
| In vivo | METHODS: To assay anti-tumor activity in vivo, Torin 1 (20 mg/kg, 20% N-mentyl-2-pyrrolidone, 40% PEG400 and 40% water) was administered intraperitoneally to NCR nude mice bearing human glioblastoma U87MG once daily for ten days. RESULTS: Continuous administration of the drug for ten days resulted in greater than 99% inhibition of tumor growth. Tumor growth continued after discontinuation of the drug, suggesting that Torin 1 treatment is primarily cytostatic and that a large number of tumor cells remain viable during treatment. [3] |
| Kinase Assay | mTORC1 and mTORC2 in Vitro Kinase Assays: To produce soluble mTORC1, HEK-293T cell lines that stably express N-terminally FLAG-tagged Raptor are generated using vesicular stomatitis virus G-pseudotyped MSCV retrovirus. For mTORC2, similar HeLa cells that stably express N-terminally FLAG-tagged Protor-1 are generated. Both complexes are purified by lysing cells in 50 mm HEPES, pH 7.4, 10 mm sodium pyrophosphate, 10 mm sodium β-glycerophosphate, 100 mm NaCl, 2 mm EDTA, 0.3% CHAPS. Cells are lysed at 4 °C for 30 min, and the insoluble fraction is removed by microcentrifugation at 13,000 rpm for 10 min. Supernatants are incubated with FLAG-M2 monoclonal antibody-agarose for 1 h and then washed three times with lysis buffer and once with lysis buffer containing a final concentration of 0.5 M NaCl. Purified mTORC1 is eluted with 100 μg/mL 3×FLAG peptide in 50 mm HEPES, pH 7.4, 100 mm NaCl. Eluate can be aliquoted and stored at -80 °C. Substrates S6K1 and Akt1 are purified. Kinase assays are performed for 20 min at 30 °C in a final volume of 20 μL consisting of the kinase buffer (25 mm HEPES, pH 7.4, 50 mm KCl, 10 mm MgCl2, 500 μm ATP) and 150 ng of inactive S6K1 or Akt1 as substrates. Reactions are stopped by the addition of 80 μL of sample buffer and boiled for 5 min. Samples are subsequently analyzed by SDS-PAGE and immunoblotting. |
| Cell Research | Cell viability is assessed with the CellTiter-Glo Luminescent Cell Viability Assay. On Day 0, 96-well plates are seeded with 500 cells per well and grown overnight. On Day 1, cells are treated with the appropriate compounds and subsequently analyzed on Days 3-5. For analysis, plates are incubated for 60 min at room temperature; 50 μL of CellTiter-Glo reagent is added to each well, and plates are mixed on an orbital shaker for 12 min. Luminescence is quantified on a standard plate luminometer. (Only for Reference) |
| Molecular Weight | 607.62 |
| Formula | C35H28F3N5O2 |
| Cas No. | 1222998-36-8 |
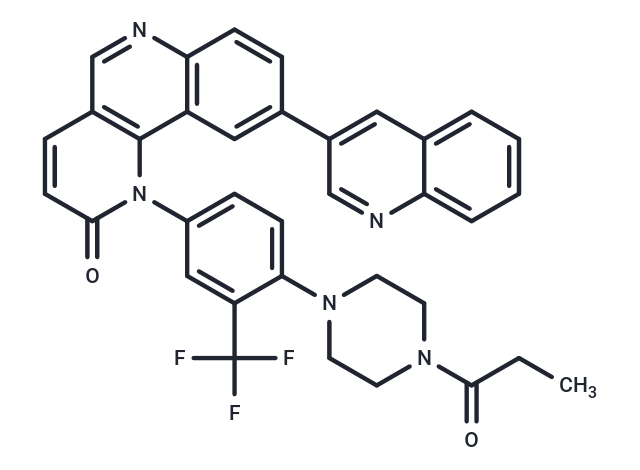
| Smiles | CCC(=O)N1CCN(CC1)c1ccc(cc1C(F)(F)F)-n1c2c(ccc1=O)cnc1ccc(cc21)-c1cnc2ccccc2c1 |
| Relative Density. | 1.362 |
| Storage | Powder: -20°C for 3 years | In solvent: -80°C for 1 year | Shipping with blue ice. |
| Solubility Information | 10% DMSO+90% Saline: 0.15 mg/mL (0.25 mM), In vivo: Please add the solvents sequentially, clarifying the solution as much as possible before adding the next one. Dissolve by heating and/or sonication if necessary. Working solution is recommended to be prepared and used immediately. DMSO: 1 mg/mL (1.65 mM), Sonication is recommended. |

Copyright © 2015-2025 TargetMol Chemicals Inc. All Rights Reserved.